Galaxy Europe has developed a pipeline to automatically create COVID-19 variant information and statistics from raw sequencing data generated by COVID-19 Genomics UK Consortium (COG-UK). The results are used to create simple and interactive visualisations so that everyone can explore this data.
Every three hours, newly published COG-UK samples are processed to produce:
- public Galaxy histories, which provide full data provenance
- variants in VCF format
- alignment files in BAM format
- consensus sequences in FASTA format
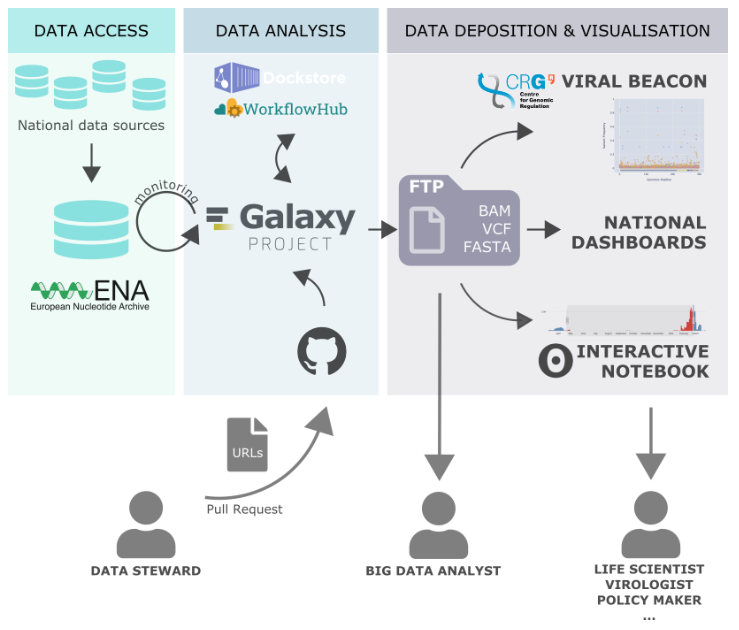
The pipeline works in cooperation with other ELIXIR-related services including workflows published by the EOSC-Life WorkFlow Hub, and makes the output of the analysis accessible through the Viral Beacon via the Centre for Genomic Regulation (CRG) at ELIXIR Spain. Full details of the process are detailed in the dedicated Galaxy Europe news release.
Alongside publishing the data for people to view and interpret, Galaxy is also openly providing the analysis pipeline so that others can replicate and adapt the analysis. Galaxy is rapidly expanding the use of this pipeline to other countries (recently integrating data from Estonia) and data repositories and is offering support to everyone who is interested in joining this effort.
Additional Galaxy COVID-19 resources
This follows from a suite of other user-friendly analysis features and pipelines that Galaxy have produced to study SARS-CoV-2. In conjunction with ELIXIR, Galaxy produced two webinar series (plus another on Advanced Features) to promote the work they have been doing on COVID-19 data analysis, the first of which was first aired in April 2020, quickly addressing the needs of researchers in the wake of the unfolding pandemic.
The above is a playlist containing the both COVID-19 webinar series recorded in 2020 and 2021 as part of the ELIXIR-Galaxy webinar series.
For further information on the ELIXIR-Galaxy Community take a look at the Community webpage.